%matplotlib inline
Process a high-resolution image
This example shows how to use squidpy.im.process with tiling.
The function can be applied to any method (e.g., smoothing, conversion
to grayscale) or layer of a high-resolution image layer of
squidpy.im.ImageContainer.
By default, squidpy.im.process processes the entire input image at
once. In the case of high-resolution tissue slides however, the images
might be too big to fit in memory and cannot be processed at once. In
that case you can use the argument chunks to tile the image in crops
of shape chunks, process each crop, and re-assemble the resulting
image. Note that you can also use squidpy.im.segment in this manner.
Note that depending on the processing function used, there might be
border effects occurring at the edges of the crops. Since Squidpy is
backed by dask, and internally chunking is done using
dask.array.map_overlap, dealing with these border effects is easy.
Just specify the depth and boundary arguments in the apply_kwargs
upon the call to squidpy.im.process. For more information, please
refer to the documentation of dask.array.map_overlap.
For the build in processing functions, [gray]{.title-ref} and
[smooth]{.title-ref}, the border effects are already automatically taken
care of, so it is not necessary to specify depth and boundary. For
squidpy.im.segment, the default depth is 30, which already takes
care of most severe border effects.
See also
examples_image_compute_smooth.examples_image_compute_gray.examples_image_compute_segment_fluo.
import numpy as np
from scipy.ndimage import gaussian_filter
import matplotlib.pyplot as plt
import squidpy as sq
Built-in processing functions
# load the H&E stained tissue image
img = sq.datasets.visium_hne_image()
We will process the image by tiling it in crops of shape
chunks = (1000, 1000).
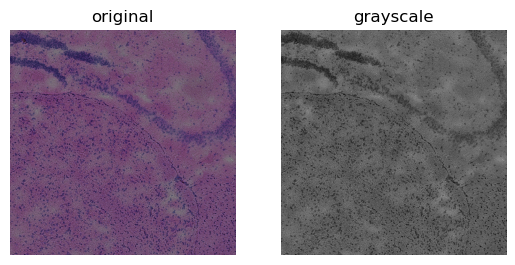
sq.im.process(img, layer="image", method="gray", chunks=1000)
Now we can look at the result on a cropped part of the image.
crop = img.crop_corner(4000, 4000, size=2000)
fig, axes = plt.subplots(1, 2)
crop.show("image", ax=axes[0])
_ = axes[0].set_title("original")
crop.show("image_gray", cmap="gray", ax=axes[1])
_ = axes[1].set_title("grayscale")
Custom processing functions
Here, we use a custom processing function (here
scipy.ndimage.gaussian_filter) with chunking to showcase the depth
and boundary arguments.
Lets use a simple image and choose the chunk size in such a way to clearly see the differences between using overlapping crops and non-overlapping crops.
arr = np.zeros((20, 20))
arr[10:] = 1
img = sq.im.ImageContainer(arr, layer="image")
# smooth the image using `depth` 0 and 1
sq.im.process(
img,
layer="image",
method=gaussian_filter,
layer_added="smooth_depth0",
chunks=10,
sigma=1,
apply_kwargs={"depth": 0},
)
sq.im.process(
img,
layer="image",
method=gaussian_filter,
layer_added="smooth_depth1",
chunks=10,
sigma=1,
apply_kwargs={"depth": 1, "boundary": "reflect"},
)
Plot the difference in results. Using overlapping blocks with
depth = 1 removes the artifacts at the borders between chunks.
fig, axes = plt.subplots(1, 3)
img.show("image", ax=axes[0])
_ = axes[0].set_title("original")
img.show("smooth_depth0", ax=axes[1])
_ = axes[1].set_title("non-overlapping crops")
img.show("smooth_depth1", ax=axes[2])
_ = axes[2].set_title("overlapping crops")
