Squidpy - Spatial Single Cell Analysis in Python
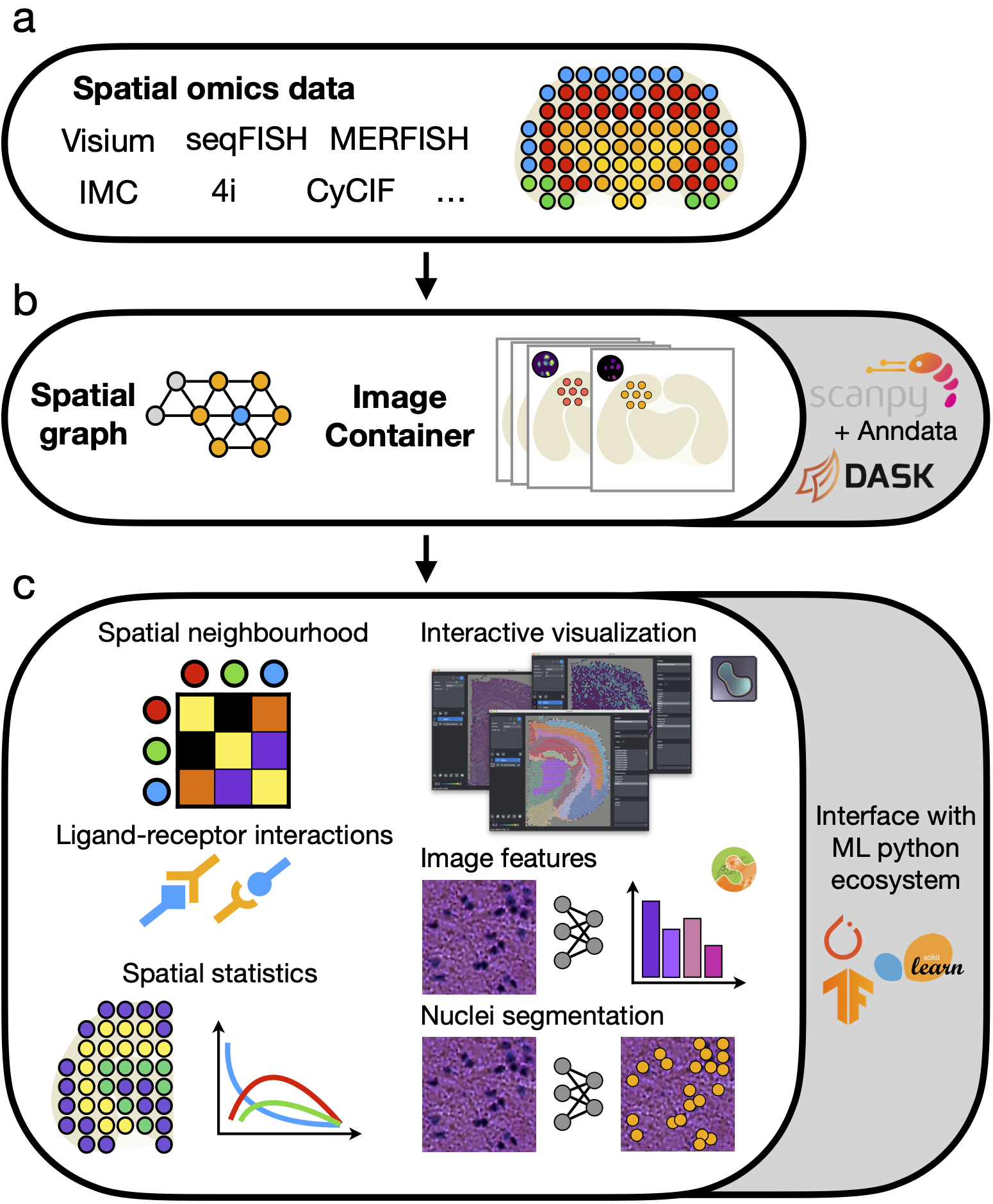
Squidpy is a tool for the analysis and visualization of spatial molecular data. It builds on top of scanpy and anndata, from which it inherits modularity and scalability. It provides analysis tools that leverages the spatial coordinates of the data, as well as tissue images if available.
Manuscript
Please see our manuscript [Palla et al., 2022] in Nature Methods to learn more.
Squidpy’s key applications
Getting started with Squidpy
Contributing to Squidpy
We are happy about any contributions! Before you start, check out our contributing guide.




