%matplotlib inline
ImageContainer object
This tutorial shows how to use squidpy.im.ImageContainer to interact with image structured data.
The ImageContainer is the central object in Squidpy containing the high resolution images.
It wraps xarray.Dataset and provides different cropping, processing, and feature extraction functions.
import numpy as np
import squidpy as sq
Initialize ImageContainer
The squidpy.im.ImageContainer constructor can read in memory
numpy.ndarray/xarray.DataArray or on-disk image files.
The ImageContainer can store multiple image layers (for example an image and a matching segmentation mask).
Images are expected to have at least a x and y dimension, with optional channel and z dimensions.
Here, we will focus on 2D images without at z dimension.
Most important arguments upon initialization are:
img- the image.layer- the name of the image layer.dims- to specify the dimensions names ofimg.lazy- set to True to allow lazy computations.scale- set this to the scaling factor between the image and the coordinates saved.
Let us see these arguments in action with a toy example.
arr = np.ones((100, 100, 3))
arr[40:60, 40:60] = [0, 0.7, 1]
print(arr.shape)
img = sq.im.ImageContainer(arr, layer="img1")
img
(100, 100, 3)
img1: y (100), x (100), z (1), channels (3)
img now contains one layer, img1. The default value of dims expects the image to have dimensions
y, x, channels or y, x, z, channels.
If the image has different dimensions, you can specify another strategy or a tuple of dimension names:
arr1 = arr.transpose(2, 0, 1)
print(arr1.shape)
img = sq.im.ImageContainer(arr1, dims=("channels", "y", "x"), layer="img1")
img
(3, 100, 100)
img1: y (100), x (100), z (1), channels (3)
Add layers to ImageContainer
You can add image layers into the ImageContainer using squidpy.im.ImageContainer.add_img().
The new layer has to share x, y (and z) dimensions with the already existing image.
It can have different channel dimensions. This is useful for add e.g., segmentation masks.
By default, unique layer and channel dimension names are chosen, you can specify them using the
layer and dims arguments.
arr_seg = np.zeros((100, 100))
arr_seg[40:60, 40:60] = 1
img.add_img(arr_seg, layer="seg1")
img
WARNING: Channel dimension cannot be aligned with an existing one, using `channels_0`
img1: y (100), x (100), z (1), channels (3)
seg1: y (100), x (100), z (1), channels_0 (1)
For convenience, you can also assign image layers directly using the new layer name:
img["seg2"] = arr_seg
img
WARNING: Channel dimension cannot be aligned with an existing one, using `channels_1`
img1: y (100), x (100), z (1), channels (3)
seg1: y (100), x (100), z (1), channels_0 (1)
seg2: y (100), x (100), z (1), channels_1 (1)
You can get a list of layers contained in an ImageContainer, and access specific image-structured arrays using their names:
print(list(img))
img["img1"]
['img1', 'seg1', 'seg2']
<xarray.DataArray 'img1' (y: 100, x: 100, z: 1, channels: 3)>
array([[[[1., 1., 1.]],
[[1., 1., 1.]],
[[1., 1., 1.]],
...,
[[1., 1., 1.]],
[[1., 1., 1.]],
[[1., 1., 1.]]],
[[[1., 1., 1.]],
[[1., 1., 1.]],
[[1., 1., 1.]],
...
[[1., 1., 1.]],
[[1., 1., 1.]],
[[1., 1., 1.]]],
[[[1., 1., 1.]],
[[1., 1., 1.]],
[[1., 1., 1.]],
...,
[[1., 1., 1.]],
[[1., 1., 1.]],
[[1., 1., 1.]]]])
Coordinates:
* z (z) <U1 '0'
Dimensions without coordinates: y, x, channelsRenaming of image layers is also possible using squidpy.im.ImageContainer.rename():
img.rename("seg2", "new-name")
img1: y (100), x (100), z (1), channels (3)
seg1: y (100), x (100), z (1), channels_0 (1)
new-name: y (100), x (100), z (1), channels_1 (1)
Visualization
Use squidpy.im.ImageContainer.show() to visualize (small) images statically.
See Show layers of the ImageContainer for more details.
For large images and for interactive visualization of squidpy.im.ImageContainer together with
spatial ‘omics data, we recommend using squidpy.im.ImageContainer.interactive(), which uses Napari.
See Interactive visualization with Napari <../tutorials/tutorial_napari.ipynb>_ for more details.
img.show(layer="img1")
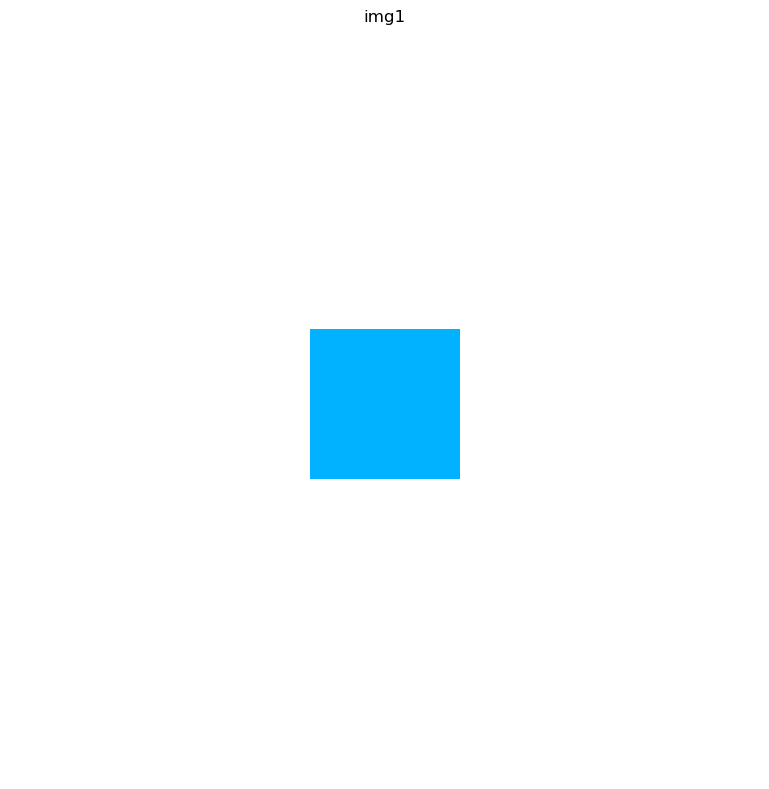
Crop and scale images
Images can be cropped and scaled using squidpy.im.ImageContainer.crop_corner() and
squidpy.im.ImageContainer.crop_center().
See Crop images with ImageContainer for more details.
crop1 = img.crop_corner(30, 40, size=(30, 30), scale=1)
crop1.show(layer="img1")
crop2 = crop1.crop_corner(0, 0, size=(40, 40), scale=0.5)
crop2.show(layer="img1")
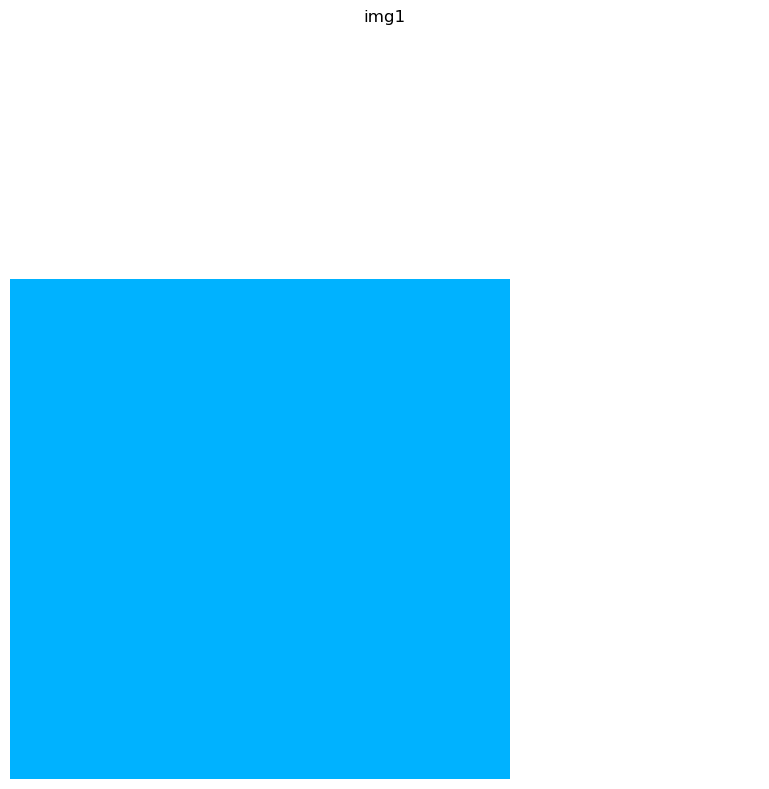
Internally, the ImageContainer keeps track of the crop coordinates in the dataset attributes.
This enables mapping from cropped ImageContainers to observations in adata for interactive
visualization and feature extraction.
Using squidpy.im.ImageContainer.uncrop(), we can reconstruct the original image.
Even when chaining multiple calls to crop, uncrop correctly places the crop in the image.
Note that uncrop only undoes the cropping, not the scaling.
print(crop1.data.attrs)
print(crop2.data.attrs)
sq.im.ImageContainer.uncrop([crop1], shape=img.shape).show(layer="img1")
sq.im.ImageContainer.uncrop([crop2], shape=(50, 50)).show(layer="img1")
{'coords': CropCoords(x0=40, y0=30, x1=70, y1=60), 'padding': CropPadding(x_pre=0, x_post=0, y_pre=0, y_post=0), 'scale': 1.0, 'mask_circle': False}
{'coords': CropCoords(x0=20.0, y0=15.0, x1=35.0, y1=30.0), 'padding': CropPadding(x_pre=0.0, x_post=5.0, y_pre=0.0, y_post=5.0), 'scale': 0.5, 'mask_circle': False}
After cropping the ImageContainer, you can subset the associated adata to the cropped image using
squidpy.im.ImageContainer.subset(). See Crop images with ImageContainer for an example.
Processing images and extracting features
The main purpose of ImageContainer is to allow efficient image processing, segmentation and features extraction.
For details on each of these steps, have a look a the following examples using the high-level API:
Process a high-resolution image for
sq.im.process().Cell-segmentation for fluorescence images for
sq.im.segment().Extract image features for
sq.im.extract_features().
These functions are build to be general and flexible. All of them allow you to pass custom processing and feature extraction functions for easy use of external packages with Squidpy.
For even more control, you can also use low-level functions provided by ImageContainer:
sq.im.ImageContainer.apply()for custom processing functions that should be applied to a specific image layer.sq.im.ImageContainner.feature_custom()for extracting features.
There are two generators, that allow you to iterate over a sequence of image crops and apply processing functions to smaller crops (e.g. to allow parallelization or processing images that won’t fit in memory:
sq.im.ImageContainer.generate_equal_crops(), for evenly decomposing the image into equally sized crops.sq.im.ImageContainer.generate_spot_crops(), for extracting image crops for each observation in the associatedadata.
Internal representation of images
Internally, the images are represented in a xarray.Dataset. You can access this dataset using
img.data.
img.data
<xarray.Dataset>
Dimensions: (z: 1, y: 100, x: 100, channels: 3, channels_0: 1, channels_1: 1)
Coordinates:
* z (z) <U1 '0'
Dimensions without coordinates: y, x, channels, channels_0, channels_1
Data variables:
img1 (y, x, z, channels) float64 1.0 1.0 1.0 1.0 ... 1.0 1.0 1.0 1.0
seg1 (y, x, z, channels_0) float64 0.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
new-name (y, x, z, channels_1) float64 0.0 0.0 0.0 0.0 ... 0.0 0.0 0.0 0.0
Attributes:
coords: CropCoords(x0=0, y0=0, x1=0, y1=0)
padding: CropPadding(x_pre=0, x_post=0, y_pre=0, y_post=0)
scale: 1.0
mask_circle: FalseWhenever possible, images are represented as lazy dask arrays.
This allows lazy computations, which only load and compute the data when it is required.
Let us load an on-disk image that is provided by the squidpy.datasets module:
By default, the lazy argument is True, therefore resulting in a dask.array.Array.
img_on_disk = sq.datasets.visium_hne_image()
print(type(img_on_disk["image"].data))
100%|██████████| 380M/380M [00:56<00:00, 7.05MB/s]
<class 'dask.array.core.Array'>
We can use squidpy.im.ImageContainer.compute() to force loading of the data:
img_on_disk.compute()
print(type(img_on_disk["image"].data))
<class 'numpy.ndarray'>
ImageContainers can be saved and loaded from a Zarr store, using squidpy.im.ImageContainer.save() and
squidpy.im.ImageContainer.load().