%matplotlib inline
Show layers of the ImageContainer
This example shows how to use squidpy.im.ImageContainer.show().
This function is useful to visualize statically different layers of the
squidpy.im.ImageContainer class.
See also
See Crop images with ImageContainer and Smooth an image for additional examples on methods of the
squidpy.im.ImageContainer.
import squidpy as sq
Load the Mibitof dataset.
adata = sq.datasets.mibitof()
We can briefly visualize the data to understand the type of images we have.
sq.pl.spatial_segment(
adata,
library_id=["point16", "point23", "point8"],
seg_cell_id="cell_id",
color="Cluster",
library_key="library_id",
title=["point16", "point23", "point8"],
)
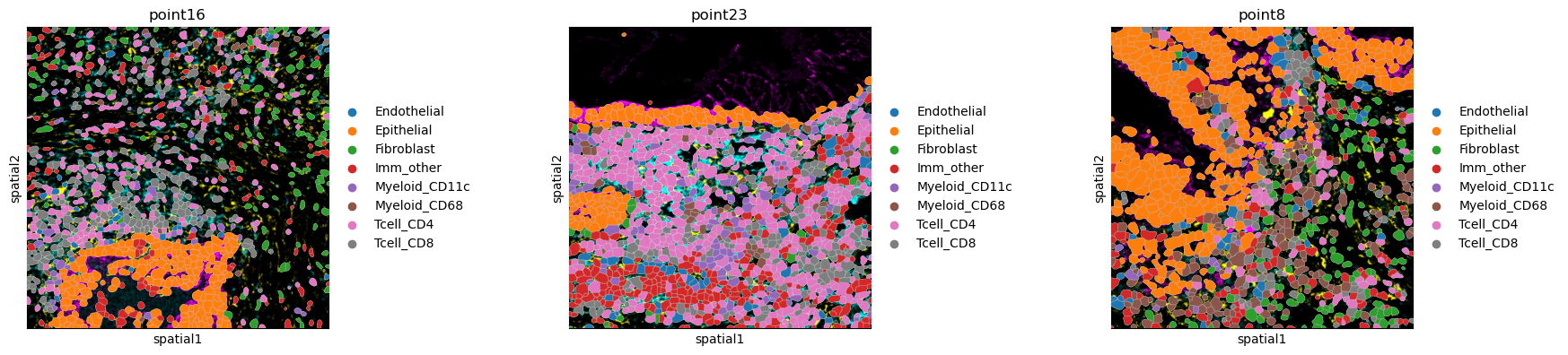
We have three different tissue samples. We also have segmentation masks
for each tissue sample. Let’s extract the image from the
anndata.AnnData object and create a squidpy.im.ImageContainer
object.
imgs = []
for library_id in adata.uns["spatial"].keys():
img = sq.im.ImageContainer(
adata.uns["spatial"][library_id]["images"]["hires"], library_id=library_id
)
img.add_img(
adata.uns["spatial"][library_id]["images"]["segmentation"],
library_id=library_id,
layer="segmentation",
)
img["segmentation"].attrs["segmentation"] = True
imgs.append(img)
img = sq.im.ImageContainer.concat(imgs)
WARNING: Channel dimension cannot be aligned with an existing one, using `channels_0`
WARNING: Channel dimension cannot be aligned with an existing one, using `channels_0`
WARNING: Channel dimension cannot be aligned with an existing one, using `channels_0`
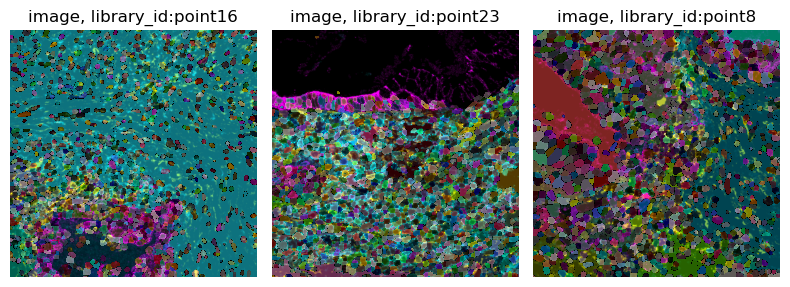
We can visualize each image of the object with
squidpy.im.ImageContainer.show().
img.show("image")
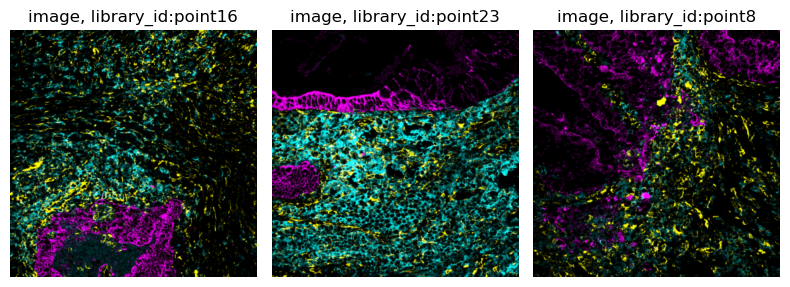
squidpy.im.ImageContainer.show() also allows to overlay the results of
segmentation.
img.show("image", segmentation_layer="segmentation", segmentation_alpha=0.5)