Nuclei segmentation using Cellpose
In this tutorial we show how we can use the anatomical segmentation algorithm Cellpose in squidpy.im.segment for nuclei segmentation.
Cellpose Stringer, Carsen, et al. (2021), (code) is a novel anatomical segmentation algorithm. To use it in this example, we need to install it first via: pip install cellpose.
To run the notebook locally, create a conda environment as conda env create -f cellpose_environment.yml using this cellpose_environment.yml, which installs Squidpy and Cellpose.
import numpy as np
import matplotlib.pyplot as plt
import squidpy as sq
Prepare custom segmentation function using Cellpose
Import the Cellpose segmentation model. See https://cellpose.readthedocs.io/en/latest/api.html#cellpose-class.
from cellpose import models
The method parameter of the sq.im.segment method accepts any callable with the signature:
numpy.ndarray (height, width, channels) -> numpy.ndarray (height, width[, channels]).
Additional model specific arguments will also be passed on.
To use the Cellpose model, we define a wrapper that initializes the model, evaluates it and returns the
segmentation masks. We can make use of Cellpose specific options by passing on arguments like the minimum
number of pixels per mask min_size.
def cellpose(img, min_size=15):
model = models.Cellpose(model_type="nuclei")
res, _, _, _ = model.eval(
img,
channels=[0, 0],
diameter=None,
min_size=min_size,
)
return res
Cell segmentation on Visium fluorescence data
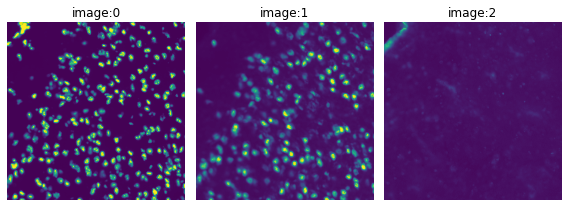
Load the image and visualize its channels.
img = sq.datasets.visium_fluo_image_crop()
crop = img.crop_corner(1000, 1000, size=1000)
crop.show(channelwise=True)
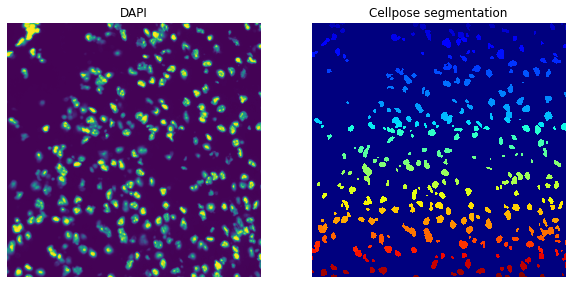
Segment the DAPI channel using the cellpose function defined above.
sq.im.segment(img=crop, layer="image", channel=0, method=cellpose)
Plot the DAPI channel of the image crop and the segmentation result.
print(crop)
print(f"Number of segments in crop: {len(np.unique(crop['segmented_custom']))}")
fig, axes = plt.subplots(1, 2, figsize=(10, 20))
crop.show("image", channel=0, ax=axes[0])
_ = axes[0].set_title("DAPI")
crop.show("segmented_custom", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("Cellpose segmentation")
ImageContainer[shape=(1000, 1000), layers=['image', 'segmented_custom']]
Number of segments in crop: 334
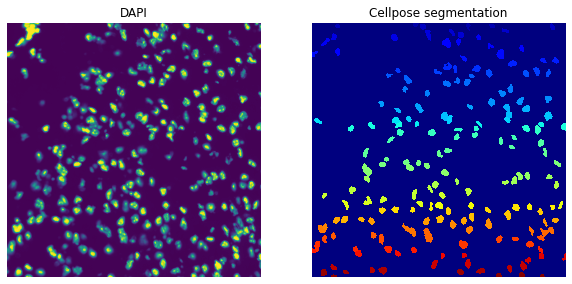
The sq.im.segment method will pass any additional arguments to the cellpose function,
so we can also filter out segments with less than 200 pixels and compare the results to the
segmentation result from above that works with the default of 15 pixels.
sq.im.segment(img=crop, layer="image", channel=0, method=cellpose, min_size=200)
print(crop)
print(f"Number of segments in crop: {len(np.unique(crop['segmented_custom']))}")
fig, axes = plt.subplots(1, 2, figsize=(10, 20))
crop.show("image", channel=0, ax=axes[0])
_ = axes[0].set_title("DAPI")
crop.show("segmented_custom", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("Cellpose segmentation")
ImageContainer[shape=(1000, 1000), layers=['image', 'segmented_custom']]
Number of segments in crop: 188
Cell segmentation on H&E stained tissue data
For the fluorescence data, we did nuclei segmentation on the DAPI channel simply by just passing on that channel to the Cellpose model. For the H&E images, we will pass on all three channels by calling sq.im.segment() with channel=None and then decide in the Cellpose model on which channel we want to segment using the first element of the channels parameter. Here, we need to be cautious as the numbering is different (0=grayscale, 1=red, 2=green, 3=blue)!
Further, for H&E images we need to invert the color values by setting invert=True.
Additionally, we found that the naive usage will recognize only few of the cells and extended the arguments we
allow for Cellpose by flow_threshold.
def cellpose_he(img, min_size=15, flow_threshold=0.4, channel_cellpose=0):
model = models.Cellpose(model_type="nuclei")
res, _, _, _ = model.eval(
img,
channels=[channel_cellpose, 0],
diameter=None,
min_size=min_size,
invert=True,
flow_threshold=flow_threshold,
)
return res
img = sq.datasets.visium_hne_image_crop()
crop = img.crop_corner(0, 0, size=1000)
crop.show("image", channelwise=True)
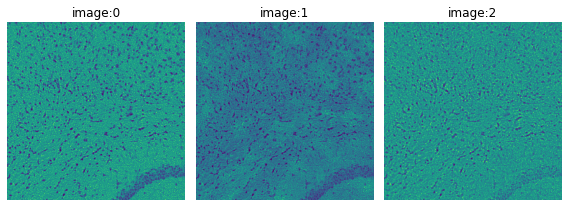
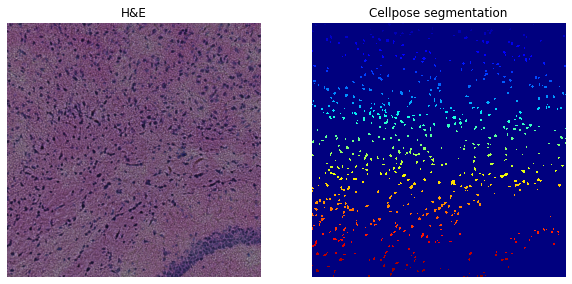
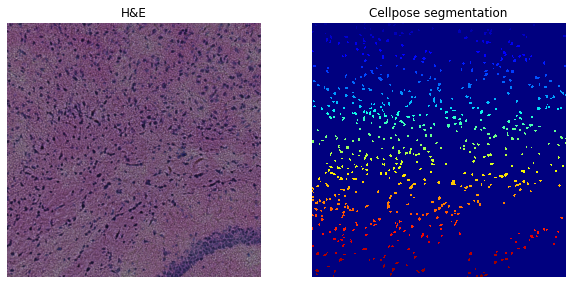
For the H&E image we start by testing the segmentation on only the blue channel (image:0).
sq.im.segment(
img=crop, layer="image", channel=None, method=cellpose_he, channel_cellpose=1
)
print(crop)
print(f"Number of segments in crop: {len(np.unique(crop['segmented_custom']))}")
fig, axes = plt.subplots(1, 2, figsize=(10, 20))
crop.show("image", channel=None, ax=axes[0])
_ = axes[0].set_title("H&E")
crop.show("segmented_custom", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("Cellpose segmentation")
ImageContainer[shape=(1000, 1000), layers=['image', 'segmented_custom']]
Number of segments in crop: 647
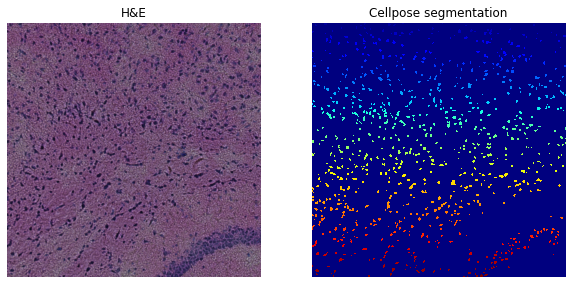
We clearly see that we should increase sensitivity and test flow_threshold=0.8 resulting in significantly more detected nuclei.
sq.im.segment(
img=crop,
layer="image",
channel=None,
method=cellpose_he,
flow_threshold=0.8,
channel_cellpose=1,
)
print(crop)
print(f"Number of segments in crop: {len(np.unique(crop['segmented_custom']))}")
fig, axes = plt.subplots(1, 2, figsize=(10, 20))
crop.show("image", channel=None, ax=axes[0])
_ = axes[0].set_title("H&E")
crop.show("segmented_custom", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("Cellpose segmentation")
ImageContainer[shape=(1000, 1000), layers=['image', 'segmented_custom']]
Number of segments in crop: 902
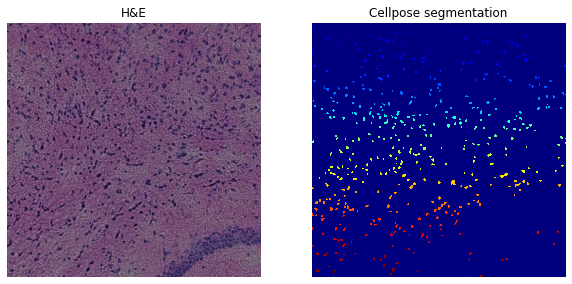
We can also check whether using all channels improves nuclei detection by segmenting on the grayscale image (channel_cellpose=0).
sq.im.segment(
img=crop, layer="image", channel=None, method=cellpose_he, channel_cellpose=0
)
print(crop)
print(f"Number of segments in crop: {len(np.unique(crop['segmented_custom']))}")
fig, axes = plt.subplots(1, 2, figsize=(10, 20))
crop.show("image", channel=None, ax=axes[0])
_ = axes[0].set_title("H&E")
crop.show("segmented_custom", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("Cellpose segmentation")
ImageContainer[shape=(1000, 1000), layers=['image', 'segmented_custom']]
Number of segments in crop: 449
And again with increased flow_threshold=0.8.
sq.im.segment(
img=crop,
layer="image",
channel=None,
method=cellpose_he,
flow_threshold=0.8,
channel_cellpose=0,
)
print(crop)
print(f"Number of segments in crop: {len(np.unique(crop['segmented_custom']))}")
fig, axes = plt.subplots(1, 2, figsize=(10, 20))
crop.show("image", channel=None, ax=axes[0])
_ = axes[0].set_title("H&E")
crop.show("segmented_custom", cmap="jet", interpolation="none", ax=axes[1])
_ = axes[1].set_title("Cellpose segmentation")
ImageContainer[shape=(1000, 1000), layers=['image', 'segmented_custom']]
Number of segments in crop: 689